so far, not so good. but to be fair we're still only running on a single machine, this stuff was made for a cluster...
running on ec2
setup
ec2 is a great way to get a bunch of machines for a bit and hadoop is well supported with scripts provided to
manage an ec2 cluster.
the first thing to do is prep the data and get it into the cloud. the complete steps then are...
- remove the prj gut header/footer with the clean text scripts, unzipped this is 2.8gb
- run rake prepare_input to reformat each file as a single line, gzipped results in 980mb
- use chunky.rb to chunk these 7,900 files into 98 files of 10mb each
- upload from home machine to s3 with s3cmd
- fire up ec2 instances
- download from s3 to hdfs using the hadoop distcp tool
- go nuts
the main pro of this approach is that it minimising the the time 'wasted' between firing the ec2 instances up and having the data in hdfs.
moving the data from s3 to hdfs is fast (an intracloud transfer) and the distcp works in parallel
a minor con is that the optimal size of file in hdfs is 64mb not 10mb but i don't want to make the smallest size 64mb since it'll be
more awkward to do smaller scale tests.
results
so how does it preform? let's use a 10 node cluster running amazons medium cpu instance.
these 10 notes provide a 30/30 map/reduce capability.
since the data size is quite small had to up the mapred.map.tasks and mapred.reduce.tasks to 30/30.
| #chunks | num docs | total tokens | unique tokens | ruby time | cluster time |
| 1 | 56 | 5.6e6 | 75e3 | 4m 32s | 10m 37s |
| 2 | 110 | 11e6 | 98e3 | 8m 48s | |
| 3 | 177 | 16e6 | 117e3 | 12m 17s | 17m 50s |
| 4 | 233 | 22e6 | 134e3 | 16m 45s | |
| 5 | 306 | 27e6 | 150e3 | 20m 15s | 24m 49s |
| 6 | 378 | 33e6 | 162e3 | 24m 13s | |
| 7 | 450 | 38e6 | 173e3 | 28m (40m) | 33m 12s |
| 8 | 531 | 44e6 | 189e3 | 32m 24s | |
| 9 | 599 | 50e6 | 197e3 | 36m 49s | |
| 10 | 686 | 55e6 | 212e3 | 41m 29s | 45m 22s |
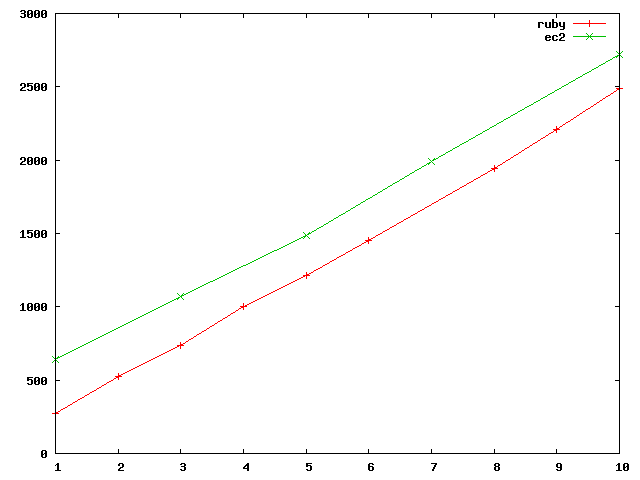
this is quite interesting...
extrapolation says hadoop will overtake the ruby one at a runtime of 2h 5min (310mb). though with a sample size this small though
such extrapolation needs to be taken with a grain of salt. so hadoop is roughly 6min + 4min per 10mb processed, ruby is 4min per 10mb.
so what conclusions can we draw? for me these results raises more questions than they answer.
- is it that hadoop is scalable but not that performant? possible since the infrastructure cost is large. i would do some longer tests on ec2 but it's not free...
- have i made a fundamental mistake regarding my map reduce implementation? are my joins terribly implemented?
- could i have implemented the algorithm using less passes of the data?
- is streaming that much slower than native java? i can't see how it would make that much of a difference
- how would pig go? especially for the expensive join steps?
hmm. some more investigation is required!
<< take 3: markov chains
index
sept 2009
